Part of this code is now incorporated in this project: https://github.com/ms-iot/plotmyface
Introduction
I recently published the article An alternative introduction to Genetic and Evolutionary Algorithms, in which I presented an Evolutionary Algorithm that's capable of finding some math formulas.
The important traits there were the creation of populations, the selection of the best results when we don't have a satisfactory result and the reproduction with mutation. Those are all traits used by the Genetic Algorithms, yet my sample was not a Genetic Algorithm because it didn't used chromosomes and genes.
Well, this time I will present a real genetic algorithm with the purpose of solving the Travelling Salesman Problem (often presented simply as TSP).
Genes and chromosomes
Maybe the most important trait to have a Genetic Algorithm is the analogy to biology that requires the use of chromosomes and, consequently, the use of genes.
Many documents say that the chromosomes are usually represented as strings and the parts of that string are the genes. The samples in those documents actually use the string data type, so each character in the string is interpreted as a gene.
In the Travelling Salesman sample I am not using the string data type. Each gene is a Location that must be visited and the chromosomes are arrays of those Locations, effectively telling the travel plan (first we go there, then there...). The size of those chromosomes is the number of locations that must be visited and, to have a valid chromosome, all the locations must be visited exactly once.
It is important to note that the use of arrays doesn't violate the idea of "strings", as that's not a data-type name, it is simply an indication that we need a sequence of genes, and arrays do that job very well.
The basic algorithm
After having a list of locations that must be visited, an initial population of chromosomes (the travel plans) needs to be generated. To do that, it is enough to randomly order those places (yet other algorithms could be used here).
After creating that initial population, the code enters a loop in which it:
- Selects the best chromosomes (the travel plans);
- Presents the best solution up to that moment;
- Reproduces them and continues the loop with the next generation.
The things that are specific to the problem are:
- How does it decide which are the best chromosomes?
- How does it decide when to stop?
- During the reproduction, will it use crossovers, mutations or what exactly to evolve?
My answers to make the sample are:
- It simply calculates the total distance traveled considering the order of the visited locations. The start location, which is the end location too and is always located in the top-left, is not encoded in the chromosomes to avoid wasting time with chromosomes that start at the wrong place, yet it is used to calculate the distance travelled;
- It doesn't decide when to stop. The algorithm continues to run forever. It is the user of the application that may consider that a good solution was found (or is simply tired of waiting) and will close the application or try with new destinations;
- Doing crossovers and random mutations will definitely create chromosomes that visit the same location twice. It may eventually evolve to the good solution but it will waste lots of time with those wrong solutions. This problem is a large one, so I will explain it in more detail.
Permutations
See the next image.
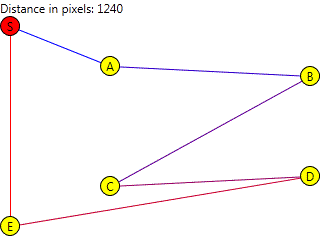
The red circle represents the start and end location. All other circles represent the locations that must be visited.
The chromosomes will actually be built of arrays of Location references which, for the article, will be represented by letters. The destinations as they were given may be seen as a base chromosome ABCDE as previously shown in the image.
When reproducing it, we don't want things like ABCBE, as we will be visiting B twice and we will never visit D. It can be represented graphically like this:

We also don't want things like ABCDF, as the F location simply doesn't exist. We only want to change the order in which the locations are visited, creating things like:
ABECD

Or
EBCDA

This is why, during the reproduction, I made the code use mutations like "swaps" and not as simply random changes of bits.
Of course that my previous examples didn't give the right result, but by doing those swaps we expect that, at some moment, it achieves this result:
ABDCE

More complex situations
Move
Now imagine that we have the following situation:

The E item is in the wrong order. We can represent this travel plan as EABDC and we must make it become an ABDCE to solve this problem, which means that we need to "move" the first item to the last position, also moving all the other items one position near the beginning. This is not achievable by using a single swap, so I decided to add such "move" operation as one of the possible mutations. In this situation, by moving the first item to the last item, we transform an EABDC into an ABDCE, effectively changing this:

into this:

Reversed range
Now, look at this problem:

We can represent this situation as AEFCBDG and, similar to the previous case, those crossed lines mean we aren't using the fastest path. We simply need to avoid those crossed lines but to do that, differently than in the previous example, the items EFCB will need to be visited in the reverse order, which is not achievable by a single swap or move.
Counting that we will have two swaps in the right place is bad, as a single swap may change the result to AECFBDG, causing this:

And the entire solution will be seen as worse, probably dying without any chance to reproduce. So, to allow the code to solve this problem in a single step I also added the support to reverse the items in a range. Surely this will happen at random and it may change AEFCBDG into CEFABDG

... yet it is possible that it corrects the chromosome, generating the ABCFEDG solution without creating worse intermediary solutions, looking like this:

Conclusion on mutations
So, the swaps, the moves and the reverse ranges are the 3 kinds of mutation that I put into the sample application with the purpose of generating better solutions in a single step. Surely those changes are made randomly and lots of times this will generate worse results, yet it is always possible to generate a better result with one of those mutations at the right place without requiring intermediate results that are seen as worse.
Elitism
Elitism is keeping some of the best solutions alive and without changes from one generation to the other. In the sample application I used it, but instead of keeping some of the best solutions alive I only keep the best one alive.
Keeping diversity
Going in the opposite direction of the Elitism, it is always good to keep diversity among the population. I didn't put any special rule to "save" different beings from dying but I created a method that forces mutation on duplicated results. Actually this kind of mutation is more useful when the crossovers aren't used.
Sample
Now, I really believe the best thing to do is to run the sample.

It allows you to increase or reduce the number of destinations, to reset the search for the best path and to create new random destinations. It also allows you to configure if crossovers are used and some other parameters that affect the "reproduction".
It doesn't allow you to draw the destinations but, well, how the destinations are generated doesn't change the search algorithm, so if you decide to change the code to allow the destinations to be drawn by users, the search algorithm will not change.
Crossovers - Another kind of problem
Most documents say that we must have a high-percentage of crossovers and only some mutations. But, when using crossovers, how do we avoid visiting the same place twice?
For example, look at this situation:
ABCDE

And EDCBA

Both alternatives are equally as good (or equally as bad, if you prefer) as they simply do the same travel in opposite directions and they can be chosen to do a crossover. When doing crossovers we will have locations visited twice (and consequently others not visited).
So, can the Travelling Salesman Problem use crossovers?
The answer is something in-between. So, explaining it, yes, it can use crossovers, as long as there's something to correct those duplicated locations.
About correcting the duplicated locations, that's something that could be used even when we accept random mutations. To make it clear, to visit ABCDE the genes could actually say AAAAA. This could be interpreted as "visit the first available location" five times.
Obviously, as soon as a location is visited it is not in the list anymore of available locations anymore, so such way of reading values will visit the locations that were named as ABCDE. If a value of F was given as the first option, well, the 6th item could be interpreted as being the same as the first when there are only 5 available locations (this means we process the list of available locations as "circular"). Then F would be the second item when there are 4 available locations. I think you got the idea. So, we could consider any value as a valid one, simply by "reading" the chromosomes differently. This will still allow terrible paths, but we will never read a "nonexistent" or "duplicated" location simply because we will interpret the strings differently.
In the sample
Well, my algorithm doesn't use characters to parse them later. In the article I am saying location A, but the code will have a reference to an actual Location instance. So, something that we represent in text as AAAAA will be really pointing to the same location 5 times.
My solution was to make the gene become valid just after doing the crossover. To do this, I create a hashset with all available locations, then I do a foreach over all the Locations in the chromosome, removing those from the list of available locations or, if that's not possible, registering that such gene must be replaced by an available one. I first finish that loop and then, if there are genes to be replaced, I simply replace them by the ones that are still available, in order. Maybe the fact that I do it in order is not the best solution, yet I avoid creating "travelling paths" that visit some locations twice and forget to visit others.
Choosing a mate
Independently of the problems related to duplicated genes for the Travelling Salesman Problem, the crossover requires two beings to mate. Here we don't see the beings, only the chromosomes, but we can consider each chromosome to have exactly one corresponding being. So, how are the beings chosen to mate?
This is an area where there are many different algorithms. In some solutions all "beings" are given a chance, which is bigger or smaller accordingly to their "fitness score". This usually makes the best beings have more children while the worst have no children, yet sometimes the best don't have a single child while some of the worst have... that's luck, really.
In my algorithm I simply ignored that. I used something very similar to what I did in the previous article. I chose the half best chromosomes to reproduce twice. Then, if they need a crossover, they will chose any of those half best chromosomes (er... beings) to reproduce. In fact my algorithm is so bad in this point that a chromosome may end-up mating with itself... creating a child that's a perfect clone, as there's nothing in the crossover to change.
Why I didn't use any of the other techniques? Simply because I think it was not needed to make this sample work. In fact, even without the crossovers the sample was already working. So, why complicate something that's already working? Well, if the purpose is to have a really complete algorithm, I may think about changing the application if I have enough requests.
Conclusion
My conclusion is that we don't need to apply or even know all the "basics" of genetic-algorithms to make a functional genetic algorithm. The sample application is capable of working and finding results by using mutations only, completely ignoring crossovers. Surely it is good to understand the other concepts and they may be necessary for professional applications, yet I believe that by building the most basic (and simpler) algorithms we can already see the algorithm working and, when we see that the algorithm is not finding the best solutions (or is too slow to find it) we can see how important all the other "details" are and we will be able to better understand them by exploring different solutions in many parts of the main algorithm.
Those many details include how to create the first population, how to chose the chromosomes that will be kept alive from one generation to another (if any), which ones will be able to reproduce, how mutations really work (I used 3 kinds of mutation in the sample), if crossovers will be used, how exactly those crossovers will work, as I chose to copy one block of genes from one chromosome to the other, but we can chose each gene randomly between the two (or more) involved chromosomes, how to calculate the fitness of the chromosomes (how good they are) etc.
So, I hope this article with its sample help new developers that want to learn the very basic of genetic algorithms. I really hope that you try the sample and even do some modifications if that helps you to better understand the concept.
 General
General  News
News  Suggestion
Suggestion  Question
Question  Bug
Bug  Answer
Answer  Joke
Joke  Praise
Praise  Rant
Rant  Admin
Admin 







 What is your assessment ?
What is your assessment ? ! Keep up the good work, and a a happy new year to you
! Keep up the good work, and a a happy new year to you