Here we’ll show you how to build a network from scratch and then train it to classify chest X-ray images into COVID-19 and Normal.
In this series of articles, we’ll apply a Deep Learning (DL) network, ResNet50, to diagnose Covid-19 in chest X-ray images. We’ll use Python’s TensorFlow library to train the neural network on a Jupyter Notebook.
The tools and libraries you’ll need for this project are:
IDE:
Libraries:
We are assuming that you are familiar with deep learning with Python and Jupyter notebooks. If you're new to Python, start with this tutorial. And if you aren't yet familiar with Jupyter, start here.
In the previous articles of this series, we used a transfer learning-based approach to fine-tune an existing ResNet50 model to diagnose COVID-19. In this article, we’ll show you how to build a network from scratch and then train it to classify chest X-ray images into COVID-19 and Normal.
Install Libraries and Load Dataset
We’ll use only TensorFlow, Keras, and OS, along with some basic additional libraries, to build our network for diagnosing COVID-19.
import tensorflow as tf
from keras import optimizers
import os, shutil
import matplotlib.pyplot as plt
First, let’s load the data that will be used to train and test the network. In this case, we’ll use a different loading technique from the one we’ve used for the transfer learning-based network. We’ll use the same dataset though.
base_dir = r'C:\Users\abdul\Desktop\ContentLab\P1\archive\COVID-19 Radiography Database’
train_dir = os.path.join(base_dir, 'train')
test_dir = os.path.join(base_dir, 'test')
train_COV19_dir = os.path.join(train_dir, 'COV19')
train_Normal_dir = os.path.join(train_dir, 'Normal')
test_VOV19_dir = os.path.join(test_dir, 'COV19')
test_Normal_dir = os.path.join(test_dir, 'Normal')
Now, let's print the numbers of the COVID-19 and Normal images in our training and testing sets.
print('total training COV19 images:', len(os.listdir(train_COV19_dir)))
print('total training Normal images:', len(os.listdir(train_Normal_dir)))
print('total test COV19 images:', len(os.listdir(test_VOV19_dir)))
print('total test Normal images:', len(os.listdir(test_Normal_dir)))
The output will be as follows:
Preprocess Data
Before feeding data to the network, images must be preprocessed. For our network-to-be-built, we’ll choose the input format of 128x128x3. All images will be rescaled to this size, which is relatively small — less computation cost. Batch size and class mode need to be set now, for ImageDataGenerator to use.
from keras.preprocessing.image import ImageDataGenerator
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(train_dir, target_size=(128, 128), batch_size=5, class_mode='categorical')
test_generator = test_datagen.flow_from_directory(test_dir, target_size=(128, 128), batch_size=5, class_mode='categorical')
To check the data and label batch shape, we will use:
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
Note that a sequential model expects an input of 4 dimensions (Batch size, X, Y, Channels). This is why the batch should be set before building your network; otherwise, an error will occur.
Build DL Network
A Keras model uses the sequential class to create layers. This is a functional model class that represents a stack of layers. It can be easily imported from Keras.
For our model, we’ll use two layers of Convolution, two of MaxPooling, three of ReLU, one Flatten, and two Dense layers (fully connected). We need to import all the required layers from Keras models such as Conv2D, MaxPooling, and Dense.
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Flatten
from tensorflow.keras.layers import Conv2D
from tensorflow.keras.layers import MaxPooling2D
from tensorflow.keras import backend as k
input_shape=(128, 128, 3)
model = Sequential()
model.add(Conv2D(32, kernel_size=(5, 5), strides=(1, 1),
activation='relu',
input_shape=input_shape))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Conv2D(64, (5, 5), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Flatten())
model.add(Dense(512, activation='relu'))
model.add(Dense(2, activation='softmax'))
model.summary()
As you can see above, a convolution layer is called using the Conv2D function, which has three main parameters:
- Filters: sets the number of filters generated in a convolution layer
- Kernel size: the size of the filter, or kernel, used in convolution (must be an odd number)
- Strides: a 2-tuple of integers that defines how the convolution will go along the X and Y axes of the input image
The pooling layer also defines stride (to go through all images) and pool _size, which is the size of the pooling filter, or kernel, used to apply pooling over the input image.
Finally, we can visualize our network:
model.summary()
Train the Network
To train our freshly built network, we need to specify the loss function and optimizer. For this network, we’ll use binary Cross-Entropy, as our target is to classify chest X-rays into two classes: Covid-19 and Normal. We’ll use Stochastic Gradient Descent as an optimizer.
In optimization functions, we need to set the learning rate. This is an important parameter. If it is set too low, the learning process may be longer because the weights will encounter small updates. On the contrary, if it is set too high, the network may overfit. It is preferable to start training our network by setting a low value of learning rate, then gradually increase it while monitoring the network performance.
In this, let's select the learning rate of 0.001.
from keras import optimizers
model.compile(loss='categorical_crossentropy',
optimizer=optimizers.adam(lr=1e-4),
metrics=['acc'])
Then, we can start training with 10 epochs (figure 11):
history = model.fit_generator(train_generator, steps_per_epoch=100, epochs=10)
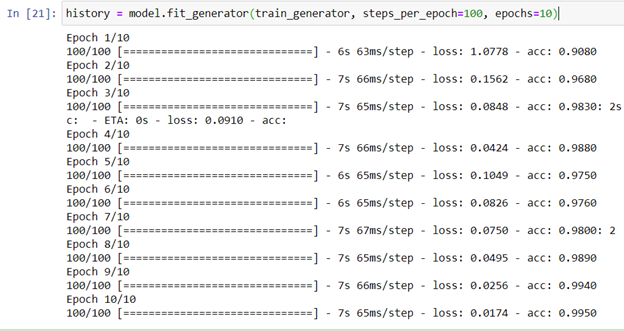
Figure 11: Training process snapshot
Network accuracy and loss during training can then be plotted using:
acc = history.history['acc']
loss = history.history['loss']
plt.figure()
plt.plot(acc, label='Training Accuracy')
plt.ylabel('Accuracy')
plt.title('Training Accuracy')
plt.figure()
plt.plot(loss, label='Training Loss')
plt.ylabel('Loss')
plt.title('Training Loss')
plt.xlabel('epoch')
plt.show()

Figure 12: Accuracy versus number of epochs
Evaluate the Network
As discussed in the previous article, the model can be tested for accuracy on new, unseen images of COVID-19 and Normal classes. In this section, Network was tested on 895 images of COVID-19 and Normal chest X-rays. We used the same testing command used in the previous pre-trained network’s testing "model.evaluate". As seen in figure 10, the network achieved a testing accuracy of 98.8% (figure 13).
Testresults = model.evaluate(test_generator)
print("test loss, test acc:", Testresults)

Figure 13: A snapshot of the testing accuracy
Next Steps?
We’ve reached the end of our series. We have achieved our set goal which was to classify chest x-rays images into either COVID-19 or Normal. Our employed models achieved well in diagnosing COVID-19 as they reached high testing accuracies of 95% and 98.8% for the transfer learning based model and the newly built model, respectively.
Although our COVID-19 classifier works well, there's still room to improve it. A key addition would be to do additional training to help the network better distinguish COVID-19 from other chest diseases such as viral pneumonia, bacterial pneumonia, mass etc.
Dr. Helwan is a machine learning and medical image analysis enthusiast.
His research interests include but not limited to Machine and deep learning in medicine, Medical computational intelligence, Biomedical image processing, and Biomedical engineering and systems.
 General
General  News
News  Suggestion
Suggestion  Question
Question  Bug
Bug  Answer
Answer  Joke
Joke  Praise
Praise  Rant
Rant  Admin
Admin